
Faculty of Agronomy – National University of Rosario, Parque Villarino, Zavalla (Santa Fe, Argentina)
Participants : (from left to rigth) Giovani Gabelli (MAD member), Julien Serret (IRD, teaching), Olivier Leblanc (MAD member), Tomás Petiti, Sol Agostina Figueroa, Alicia Zelada, Celeste Arancibia, Florencia Rocío Turco, Larisa Altube, Liliana Alvarez, Mauro Debetti, Sofía Gischuck, Pilar Alda, Augusto Villalba (MAD member), François Sabot (IRD, teaching), Ingrid Georgina Orce, Julie Orjuela (IRD, teaching), Davide Perrone (MAD member), Francisco De Blas, Martín Raúl Quevedo (MAD member), Marie Couderc (IRD, teaching)
Objective: The aim of the course was to train plant biologists (mostly PhD students and post-docs) in Oxford Nanopore genomic sequencing and in sequence analysis. It consisted of practical laboratory (HMW DNA extraction, library preparation, sequencing) and bioinformatics (base calling, read quality and filtering, assembly) sessions, as well as of lectures.
Organizers: Olivier Leblanc (IRD, MAD coordinator) & Juan Pablo Ortiz (IICAR-CONICET-UNR)
Local organizers: Maricel Podio & Juliana Stein (FCA-UNR)
Lab team: Marie Couderc (IRD), Lorena Siena (IICAR), Carolina Colono (IICAR), & Julien Serret (IRD)
Bioinformatics team: Julie Orjuela (IRD), Maricel Podio (IICAR) & François Sabot (IRD)
Day one – Introduction, HMW gDNA extraction, Library preparation and loading



Laboratory teachers, Maricel Podio (IICAR), Lorena Siena (IICAR), Marie Couderc (IRD), Julien Serret (IRD)


Lab work – HMW gDNA extraction



Day 2 – Practical DNA library preparation


The 7:30 a.m. shuttle from Rosario to Zavalla and Juan Pablo’s explanations. Special thanks to our drivers, Manuel and Roberto!
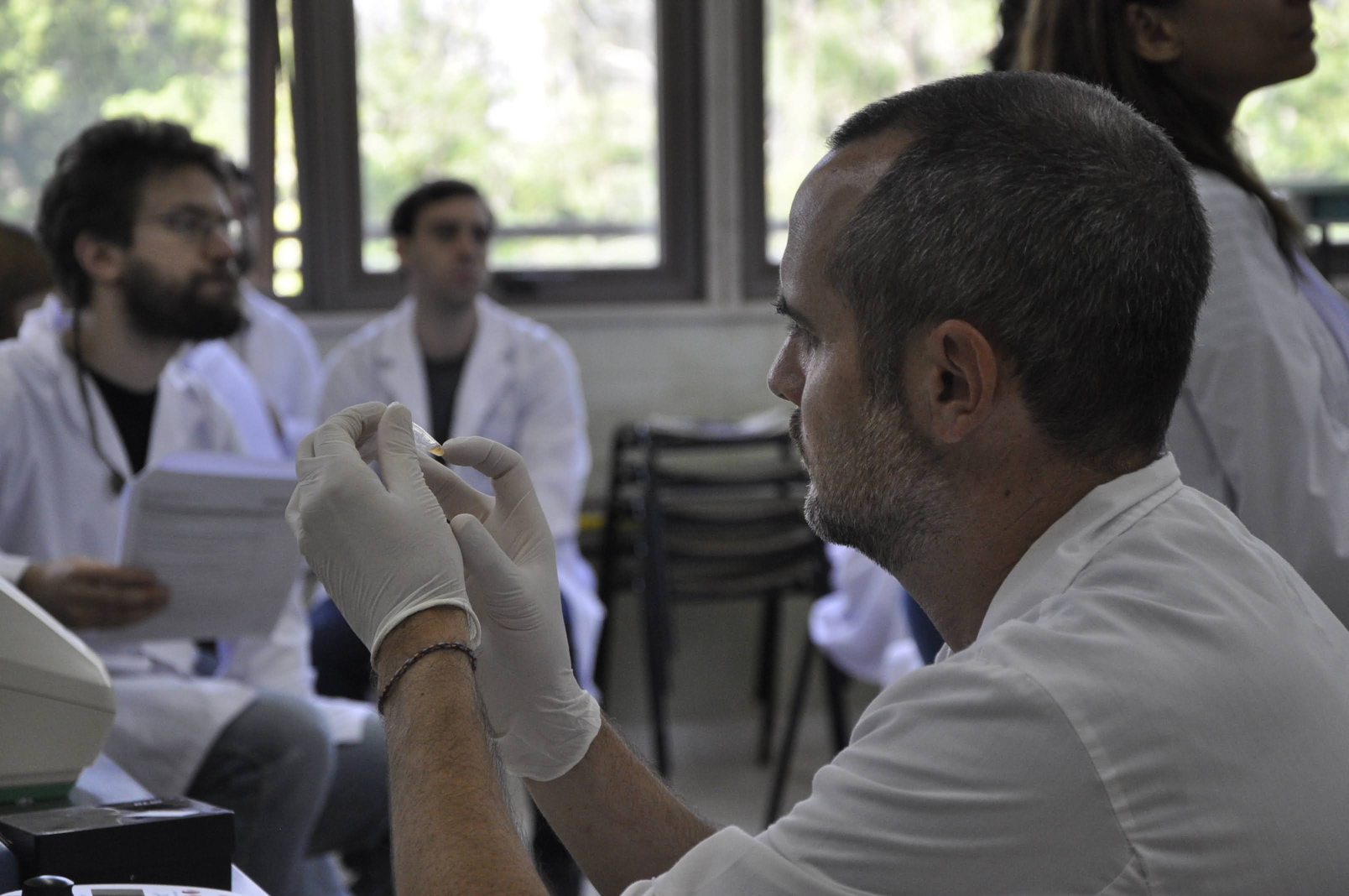
Lab work – ONT library preparation – Demo by Julien

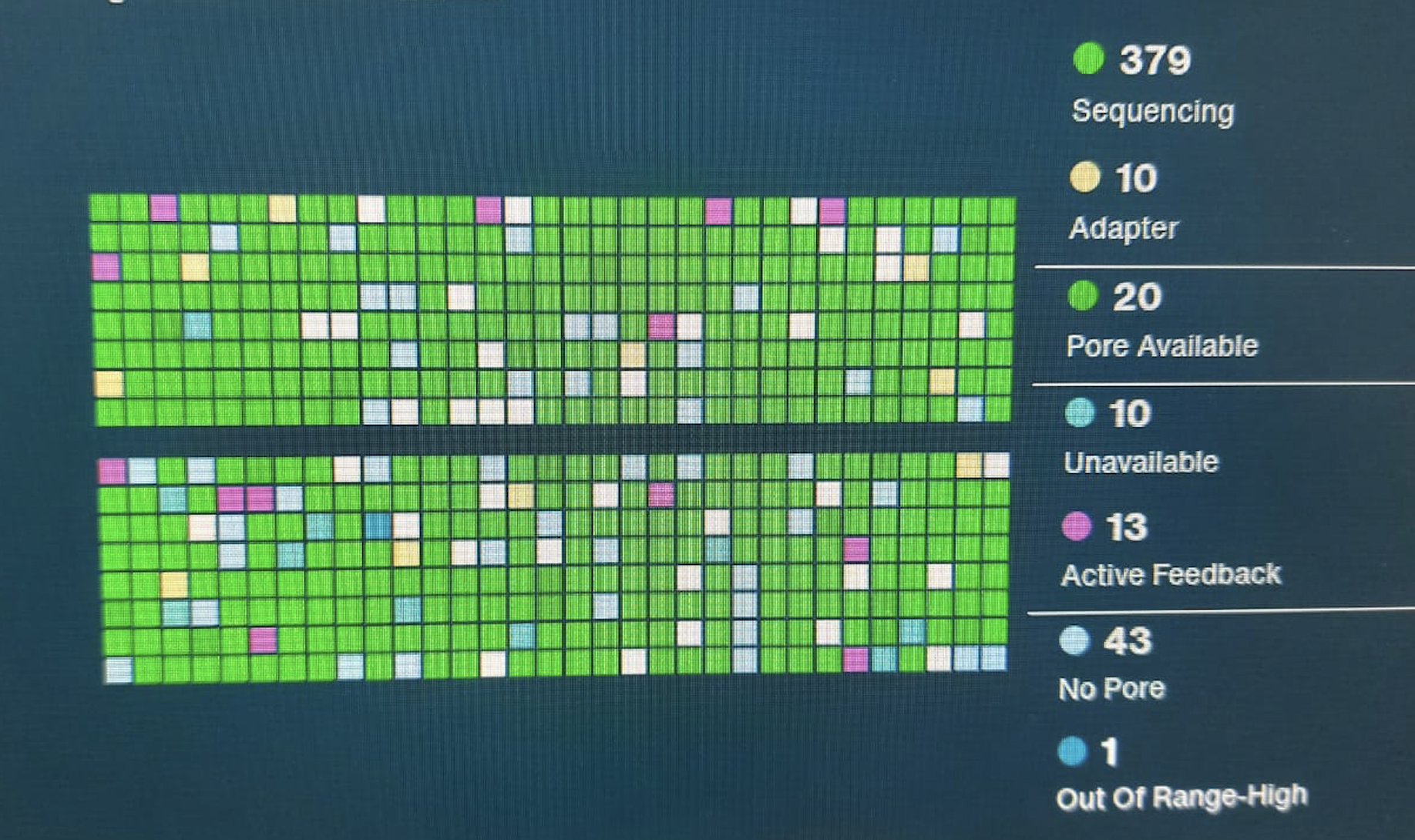
Nice flow cell!
Day 3 – Bioinformatics (base calling, quality control)


Time to join Linux!…
Does maté really boost bioinformatics skills?

Bioinformatics teachers, Julie Orjuela and Françcois Sabot, discussing the N50 metrics

Day 4 – Bioinformatics (genome assembly)



Day 5 – Wrap-up & closing


Thanks again to the IICAR and IRD people involved in training and organization. Glad to see you all wearing your guardapolvo!
Carolina, Maricel, Marie, Julien, Francois, Lorena, Olivier, Julie, and Juan Pablo
Participants
Alda Pilar, Researcher, CERZOS (CONICET-UNS), Bahia Blanca, Argentina
Altube Larisa V, Researcher, IUNIR, Rosario, Argentina
Arancibia Celeste, Post doc, IBAM (CONICET-UNCUYO), Mendoza, Argentina
Alvarez, Liliana
Bedetti Mauro, PhD student , IICAR (CONICET-UNR), Rosario, Argentina
Bianchi Julieta, Researcher, ICAR (CONICET-UNR), Rosario, Argentina
De Blas Francisco, Researcher, IBONE (CONICET-UNNE), Corrientes, Argentina
Figueroa Sol Agostina, PhD student, IPROBYQ (CONICET-UNR) Rosario, Argentina
Gabelli Giovanni, Post doc, University of Padova, Italy
Gischuck Sofía, PhD student, UNCo, Neuquén, Argentina
Godoy Federico, PhD student, IICAR (CONICET-UNR), Rosario, Argentina
Orce Ingrid Georgina, Researcher , CREAS (CONICET-UNCA), Catamarca, Argentina
Petiti Tomás, PhD student, IPROBYQ (CONICET-UNR) Rosario, Argentina
Perrone Dávide, PhD Student, University of Milan, Italy
Turco Florencia Rocío, PhD student, UNC, Córdoba, Argentina
Quevedo Martín Raúl, PhD student, CERZOS (CONICET-UNS), Bahia Blanca, Argentina
Villalba Augusto Ivan, PhD student, IBONE (CONICET-UNNE), Corrientes, Argentina
Vega María Sol, PhD student, IICAR (CONICET-UNR), Rosario, Argentina
Vega Juan Manuel, PhD student, IICAR (CONICET-UNR), Rosario, Argentina
Zelada Alicia, Researcher, UBA, Buenos Aires, Argentina


Recent Comments