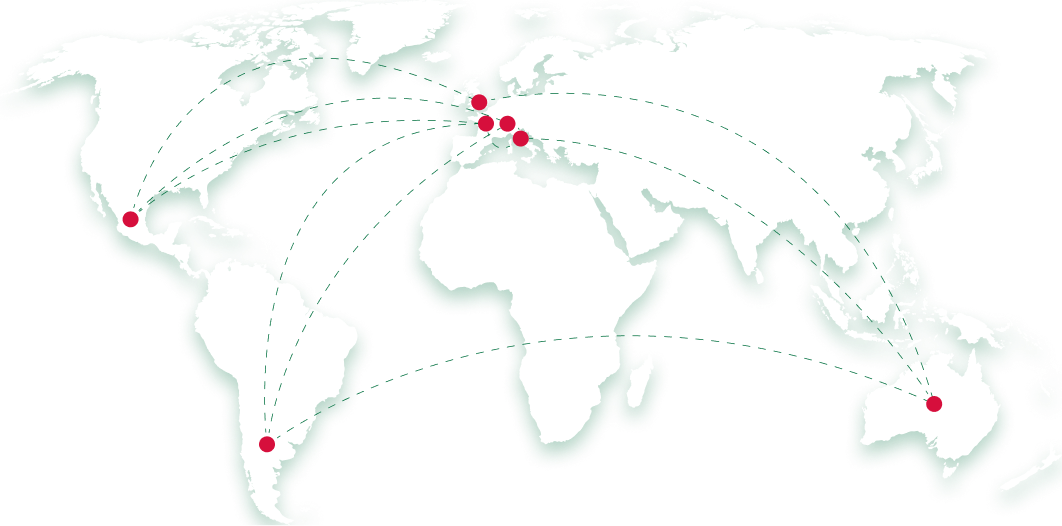
secondees
Secondment period means the period spent by the staff member in a host organisation for the purposes of the action. A secondee is a staff member hosted.
2025
Dr. Fulvio Pupilli
IBBR, Perugia, Italy
IICAR (Rosario, Argentina) from October 27, 2025 to November 28, 2025

Dr. Viviana Echenique
CERZOS, Bahia Blanca, Argentina
University of Perugia (Italy) from October 1, 2025 to October 30, 2025
Photo: Viviana Echenique at UNIPG

Juan Manuel Vega, PhD student
IICAR, Rosario, Argentina
IBBR (Perugia, Italy) from September 11, 2025 to November 12, 2025
Juan Manuel Vega, from the DREP group at IICAR-CONICET (Rosario, Argentina), will be seconded to IBBR-CNR (Perugia, Italy) to work with Dr Fulvio Pupilli’s group from 11 September to 12 November 2025. During his stay, he will work on improving a preliminary assembly of the Paspalum simplex genome by constructing a scaffold based on the P. notatum reference genome. He will also contribute to the annotation of the sexual and apomictic genomes of P. simplex and to the identification and characterisation of the apomixis control locus (ACL) by following the strategies previously developed for P. notatum. Comparative analyses will be conducted to evaluate genome structure in Paspalum species, and transcriptomic datasets from inflorescences will be used to explore gene expression in the ACL region. The work will be carried out in collaboration with Davide Perrone, Andrea Rubini and Fulvio Pupilli.
Picture: from left to right, Fulvio Pupilli, Juan Manuel Vega & Davide Perrone

Dr. Juan Pablo A. Ortiz
IICAR, Rosario, Argentina
IRD (Montpellier, France) from September 1, 2025 to October 30, 2025
As part of WP3 and 4, Juan Pablo A. Ortiz from IICAR-CONICET, Argentina, visited IRD, Montpellier, France, from September 1 to October 30, 2025. During his visit, Juan Pablo worked on the analysis of the transmission of apospory (a key component of apomictic reproduction) through female gametes. Using segregation data generated from a highly sexual facultative apomictic plant, a model for apospory inheritance by female gametes is being elaborated. The work plan also included the determination of hormones content (auxin, cytokinin, zeatin and jasmonic acid) in the reproductive tissues of sexual and apomictic Paspalum notatum genotypes. Biochemical assays were carried out at IRD by Virginie Vaissayre using the UHPLC-MS/MS method. Analysis of hormone concentration and their biological role in apomictic reproduction will be conducted in conjunction with Dr. Olivier Leblanc.
Picture: Confocal microscope at IRD, Lorena Siena (left) and Juan Pablo Ortz (right)

Dr. Maricel Podio
IICAR, Rosario, Argentina
IRD (Montpellier, France) from September 1, 2025 to October 31, 2025
Dr. Maricel Podio from IICAR (Zavalla, Argentina) performed a research secondment at IRD in Montpellier, France, from September 1 to October 31, 2025. During this period, she investigated the epigenetic landscape of Paspalum notatum by sequencing the genomic DNA extracted from the ovaries of tetraploid sexual and apomictic plants, using Oxford Nanopore technology. To initiate the characterization of their epigenomes, ovaries at the anthesis stage were selected from genotypes exhibiting different levels of apospory expressivity. The sequencing performed during the secondment will enable the identification and quantification of DNA methylation marks (5mC and 5hmC), which will then be compared with leaf DNA methylation profiles to provide insight into the epigenetic patterns associated with the mode of reproduction. All experiments were carried out in collaboration with two members of the DIADE research unit, Julien Serret and Cédric Mariac.
Picture: Lab work at IRD

Sol Vega, PhD student
IICAR, Rosario, Argentina
IRD (Montpellier, France) from September 1, 2025 to November 30, 2025
María Sol Vega is visiting IRD to further characterize the auxin-response repressor IAA16 in the model species Arabidopsis thaliana. The orthologue of IAA16, IAA30, is downregulated in the apomictic ovules of Paspalum notatum compared to sexual ones. The research activities will include determination of spatial expression pattern of IAA16 in reproductive tissue using a construct carrying the IAA16 gene fused with the Ruby reporter gene (pIAA16:IAA16:Ruby). Double homozygous plants will also be obtained and studied, derived from crosses between iaa16 and gametogenesis and embryogenesis cellular identity lines (pWOX2:CENH3, pKNU, FGR7.0, pDRN), auxin patterning lines (DR5, R2D2, PIN1, aux1), and mutant lines for its correpresor TOPLESS and the apomixis related gene TGS1. Phenotypic analysis will be carried out using DIC cytoembryology and cell type-specific reporter lines analysis under epifluorescent and confocal microscopy. Finally, María Sol will present her work at the 5th International Conference on Apomixis, which will be held in Montpellier from 16 to 19 September. The title of her presentation is “Auxin response repressor IAA16 defective mutants show developmental alterations in female gametophytes and embryos in Arabidopsis thaliana”.
Picture: Maria Sol Vega (left) & Lorena Siena (right) at IRD, Montpellier (France).
Dr. José Carballo
CERZOS, Bahia Blanca, Argentina
University of Perugia (Italy) from August 9, 2025 to October 10, 2025
Dr. Juan Pablo Selva
CERZOS, Bahia Blanca, Argentina
NIAB (United Kingdom) from July 28, 2025 to September 27, 2025

Dr. Lorena Siena
IICAR, Rosario, Argentina
IRD (Montpellier, France) from June 29, 2025 to November 30, 2025
In this secondment, Lorena Siena will advance the functional characterization of the TRIMETHYLGUANOSINE SYNTHASE1 (TGS1) and QUI-GON JINN (QGJ) genes during Arabidopsis thaliana reproductive development. The activities will be carried out in collaboration with members of the DIADE group and will including: single cell RNA sequencing using the long read ONT technology to analyze gene expression differences between Col and tgs1 A. thaliana plants and the construction of plasmids designed to study the function of TGS1; the characterization of the cellular identity of the extra functional megaspores observed in qgj ovules, particularly using the pWOX2-CENH3-GFP reporter, and; the study of the potential interactions between QGJ and TGS1 by determining the QGJ expression pattern (pQGJ-GUS) in tgs1 ovules and, reciprocally, that of TGS1 (pTGS1:TGS1-GUS) in qgj ovules. In addition, Lorena will participate of the Apomixis2025 international conference, which will be held 16–19 September in Montpellier, where she was invited to present her work on the characterization of QUI-GON JINN during female reproduction in plants.
Picture: Lorena, Bambouseraie d’Anduze, France.

Dr. Luciana Delgado
IICAR, Rosario, Argentina
IRD (Montpellier, France) from June 13, 2025 to October 10, 2025
As the final WP6 secondment for the MAD project, in collaboration with Daphné Autran, we will complete the quantitative data analysis of cell division and cell growth in Paspalum rufum ovule primordia. Our aim is to analyse the dynamics of cell growth and division in sexual and apomictic contexts to identify differential events that predict the differentiation of aposporous initials in cells surrounding the sexual germline in an apomictic grass. This will also give a comprehensive understanding of how the architecture and shape of the ovule influence the processes of cell differentiation during germline formation. We will also present and discuss our recent findings at the V International Conference on Apomixis, held in September 2025 in Montpellier, to celebrate the end of this fruitful project.
Picture: Luciana in front of the IRD greenhouses

Dr. Michele Bellucci
IBBR, Perugia, Italy
IICAR (Rosario, Argentina) from April 29, 2025 to May 30, 2025
On April 29, 2025, Dr. Michele Bellucci from CNR-IBBR, Perugia, Italy, started a secondment at IICAR until May 30, 2025. He will work on the transformation of Paspalum notatum by particle bombardment, to knock down the expression of the PnIAA30 gene in tetraploid sexual and apomictic plants using RNAi. The silencing vector was assembled at CNR-IBBR and contains a region of the PnIAA30 gene cloned in opposite orientation and the bar gene as a selectable marker. PnIAA30 was found to be downregulated in apomictic ovules compared to the sexual ovules. Our hypothesis is that PnIAA30 silencing (together with other of its paralogs) could trigger the development of parthenogenetic embryos in sexual plants and/or increase the rate of parthenogenesis in apomictic genotypes. All experiments will be carried out with Dr. Carolina M. Colono and Dr. Juan Pablo A. Ortiz.
Picture: Carolina Colono, IICAR (left) & Michele Bellucci, IBBR (right)
Dr. Fulvio Pupilli
IBBR, Perugia, Italy
IICAR (Rosario, Argentina) from April 05, 2025 to May 28, 2025

Dr. Alessandro Camperio
University of Milan, Italy
CERZOS (Bahia Blanca, Argentina) from March 17, 2025 to April 15, 2025
During this secondment, I was trained in performing bioinformatic analyses on a set of single-nucleus RNA sequencing data generated from Oryza sativa ovaries (sampled at different stages of development) under the supervision of Dr. José Carballo. The aim was to acquire knowledge in single-nucleus transcriptome analysis and to study the expression of selected ARF genes, identified in other species as putative genes involved in apospory and diplospory. As part of this work, I sought to generate high-resolution transcriptomic profiles capturing the gene expression dynamics of rapidly evolving gametophyte and sporophyte cell populations during ovule development. These single-nucleus transcriptomic data will be used for the broader purpose of comparing them with other datasets from Paspalum and Eragrostis species. This comparative framework was designed to better understand the molecular landscape discovered in rice, with a particular focus on detecting genes expressed in the nucellus and related tissues.
Picture (from left to right): Viviana Echenique, Alessandro Mariani & Alessandro Camperio
Granpierro Marconi, Msc
University of Perugia, Italy
University of Adelaid (Australia) from January 05, 2025 to March 21st, 2025
2024
Dr. Alice Casati
University of Milan, Italy
Unidad de Genómica Avanzada (Cinvestav, Mexico) from November 25, 2024 to February 15, 2025

Dr. Alessandro Mariani
University of Perugia, Italy
During my secondment at CERZOS-CONICET in Bahía Blanca (Nov 2024 – Apr 2025), I contributed to the Mechanisms of Apomictic Development (MAD) project by investigating the genetic and epigenetic regulation of apomixis in plants. My research focused on mapping loci associated with apomictic traits through QTL analysis in segregating populations, identifying candidate genes involved in diplospory, parthenogenesis, and autonomous endosperm development. I conducted bioinformatic analysis of high-throughput genotyping and sequencing data, including population structure and association studies, to uncover genomic regions linked to apomixis. Flow cytometry was employed to assess ploidy levels and genome size, supporting the phenotypic classification of accessions. Additionally, I integrated DNA methylation and RNA-seq data to detect epigenetic differences between apomictic and sexual individuals. These integrative approaches enhanced our understanding of the molecular basis of apomixis, aligning with the project’s goal to harness this trait for crop improvement.
Picture: Viviana Echenique (left) & Alessandro Mariani (right) at CERZOS.
CERZOS (Bahia Blanca, Argentina) from November 13, 2024 to May 26, 2025

Giacomo Bongiorno, PhD student
University of Perugia, Italy
CERZOS (Bahia Blanca, Argentina) from September 20, 2024 to November 21, 2024
During this secondment, I performed Presence-Absence a compative analysis of Presence-Absence Variations (PAVs) in Eragrostis curvula, leading to the identification of 503 candidate genes for apomixis. A subset of these was validated by RT-PCR. I also conducted Short-Variant (SNPs and InDels) calling, by incorporating new genotypes to further investigate genomic variation, reproductive modes, and ploidy levels. The phylogenetic tree constructed using short variants classified genotypes into two main branches, distinguishing high-polyploid from diploid and tetraploid genotypes. This analysis provided deeper insights into the genetic relationships between genotypes of different ploidy levels, shedding light on new scenarios related to genome evolution in E. curvula.
Picture: Giacomo Bongiorno (left) and José Carballo (right)

Eng. Caroline Michaud
IRD, Montpellier, France
IICAR (Rosario, Argentina) from November 12, 2024 to December 13, 2024
During this secondment, I contributed to the work of Luciana Delgado and Daphné Autran on Paspalum rufum, exploring the links between ovule primordium architecture and cell fate plasticity leading to apospory. To determine the patterns of cell proliferation during ovule development in sexual and apomictic Paspalum rufum at pre-meiotic stages, we conducted EdU labeling experiments to mark cells in S-phase of the cell cycle using the Click-iT™ EdU kit. Ovaries were dissected at four early stages, processed for EdU incorporation and detection, then imaged using a ZEISS 880 confocal microscope at the Institute of Molecular and Cellular Biology (IBR-CONICET) in Rosario. These images will be segmented with MorphographX (MGX) to quantify the differential proliferation rates between genotypes. I have also collaborated on a project by Olivier Leblanc and Lorena Siena, constructing plasmids for the functional analysis of the TGS1 gene in Arabidopsis thaliana, with completion planned in Montpellier in 2025.
Picture. Top left: my UNR teaching credential card!; Bottom left: the IBR confocal microscope, and; Right: a diagram of the dissected Paspalum rufum stages.

Dr. Olivier Leblanc
IRD, Montpellier, France
IICAR (Rosario, Argentina) from November 12, 2024 to December 13, 2024
Picture: cruzing the Pampa from Rosario to Bahia Blanca
Dr. Tamunonengiye-ofori Lawson
Cambridge, UK, NIAB
CERZOS (Argentina), from October 31, 2024 to December 15, 2024

Dr. Daphné Autran
IRD, Montpellier, France
IICAR (Rosario, Argentina) from October 28, 2024 to December 31, 2024
During this secondment, in the frame of WP6 of the MAD project, Daphné Autran (IRD) collaborated with Luciana Delgado (IICAR-CONICET), with the help of Caroline Michaud (IRD), to decipher ovule morphogenesis in the aposporous specie Paspalum rufum. To determine which are the earliest cellular events associated with a facultative aposporous developmental program, we took advantage of the flowering period of the unique population of contrasting P. rufum plants available at IICAR, Zavalla, Rosario, Argentina. Here, the process of cell proliferation was specifically addressed, by monitoring the number and localization of dividing cells, using living ovules, collected at different developmental stages. After fixation, clearing and staining, the samples were imaged using confocal fluorescence microscopy (available at IBR (Instituto de Biologia de Rosario). The 3D images dataset generated will be next analyzed, to complete the information gathered from the 3D cell growth atlas of P. rufum ovules produced in previous secondments.
Picture: Luciana Delgado (left panel) & Luciana Delgado and Daphné Autran (right panels)

Rosario, Argentina, IICAR
Dr. Silvina Pessino
University of Milan from October 2nd 2024 to November 27, 2024
Dr. Silvina Pessino (IICAR-CONICET, Argentina) was seconded to UMIL, Italy, from October 2nd to November 27th, 2024, to investigate the role of several AUXIN RESPONSE FACTORS (ARFs) and their respective miRNA controllers in the specification of the female germ line in Arabidopsis thaliana, in collaboration with Dr. Lucia Colombo and Dr. Marta Mendes. In particular, during this secondment, she participated in the final publication steps of the article Pessino SC et al. (2024) AUXIN RESPONSIVE FACTOR 10 insensitive to miR160 regulation induces apospory-like phenotypes in Arabidopsis, iScience; prepared several MAD project deliverables, and; analyzed McSeED and RNA-seq data pointing to the control of ARF expression mediated by the RdDM system.
Picture: Marta Mendes (left) & Silvina Pessino (right)

Dr. Juan Pablo A. Ortiz
Rosario, Argentina, IICAR
University of Padova from November 1st, 2024 to December 1st, 2024
Dr. Juan Pablo A. Ortiz will undertake a new secondment to work on the analysis of apomixis-related sequences, with a particular focus on the discovery of genes associated with parthenogenesis. This work will include the analysis of transcriptome databases from sexual and apomictic genotypes to identify genes related to hormone cascades, RNA processing and cell signaling that may be involved in parthenogenesis. The analysis will also include the available apomixis controlling locus genomic resources. This work will be carried out in collaboration with Dr Giovanni Gabelli of the DAFNAE group. The results obtained will contribute to WP3 and WP4 and may provide new candidates for the induction of parthenogenesis in natural apomictic species.
Picture: Sharing mate… from left to right: Juan Pablo Ortiz, Lorena Siena & Samela Draga

Dr. Lorena Siena
Rosario, Argentina, IICAR
IBBR, from August 30, 2024 to September 29, 2024
During her visit at the laboratory of Dr. Fulvio Pupilli, Lorena will sequence tetraploid sexual and apomictic plants of Paspalum simplex in order to improve current tetraploid genome assemblies. The information produced will contribute to WP3 and will be used to study the genome structure and evolution and to decipher the gene content of the apomixis locus. This work will be carried out in collaboration with Dr. Andrea Rubini and BCs Davide Perrone from IBBR and Dr Juan Pablo A Ortiz from IICAR. Lorena will also set up in situ hybridization assays to characterize the expression pattern of the P. simplex candidate gene ARF5 in apomictic and sexual ovaries. She will be involved in the design and labelling of the probe, and in the paraffin mounting of the ovaries. The sectioning and hybridization will be carried out at IICAR, CONICET-UNR. These experiments will be performed in collaboration with Dra Eugenia Cáceres from IBBR.
Picture: Lorena Siena (right) & Davide Perrone (left)

Dr. Juan Pablo A. Ortiz
Rosario, Argentina, IICAR
IBBR, from August 30, 2024 to September 29, 2024
The purpose of this secondment is to deepen the characterization of sexual and apomictic genomes of Paspalum simplex. The work plan includes DNA extractions, library preparation and ONT sequencing experiments of tetraploid plants. These activities will contribute to WP3, which aims to analyze the structure and evolution of apomictic genomes and elucidate the gene content and function of the apomixis locus. The other members of the team that will participate in this work are Dra Lorena Siena (IICAR), Dr Andrea Rubini (IBBR) and BSc Davide Perrone (IBBR). Juan Pablo will also work on the construction of an RNAi vector designed to down-regulate the expression of ORC3, one candidate gene identified by Dr Pupilli’s group as a regulator of the endosperm development in apomictic seeds. This construction will be used to generate transgenic maize plants in order to validate its function in the control of maternal genomic excess in the endosperm of a sexual crop. This part of the work will be carried out in collaboration with Dra Francesca De Marchis and Dr Michelle Bellucci, from IBBR.
Picture: Juan Pablo A. Ortiz (left) & Fulvio Pupilli (right)

Ing. David Lopez Villegas
CERZOS, Bahia Blanca, Argentina
IRD (France), from June 20, 2024 to August 26, 2024
Ing. David Lopez Villegas performed a secondment at IRD in Montpellier, France, from June 24 to August 23, 2024. During the secondment, he acquired the competencies and knowledge required to perform technology valorization in collaboration with the IRD Technology Transfer Office, with a specific focus on apomixis and its potential applications in agriculture. He conducted a systematic review of patents related to apomixis and associated technologies, providing valuable insights into current technological trends, intellectual property strategies, and the leading institutions and companies active in this field.
Picture: David Lopez at IRD, Montpellier, France.
Dr. Andrés Bellido
CERZOS, Bahia Blanca, Argentina
IRD (France), from may 31, 2024 to June 29, 2024
Arabidopsis thaliana transgenic plants were generated expressing apomixis-related genes identified in Eragrostis curvula. These plants showed several phenotypes at MMC specification and during embryo sac stages linked to apomixis, and some evidences of loss in DNA methylation. The aim of this work was to monitor the most relevant histone marks during ovule development in these Arabidopsis transgenic lines using immunolabelling and confocal microscopy techniques.

Dr. Viviana Echenique
CERZOS, Bahia Blanca, Argentina
University of Perugia (Italy), from May 30, 2024 to August 2, 2024
The aim of this secondment at UNIPG is to continue the work with Dr. Albertini´s group to identify the genomic region involved in apomixis in Eragrostis curvula and characterize its gene content. To achieve this, we will analyze the data previously obtained from the E. curvula pangenome, which consits of Illumina sequencing of 10 genotypes with similar genomic backgrounds but with different reproductive modes (apomictic and sexual) and ploidy levels (2x, 4x, 6x, 7x and 8x). The data will be used to study the genomic regions involved in apomixis and syntenic relationships with sexual counterparts through a mapping approach to obtain the first E. curvula pangenome assembly.

Dr. Maria Cielo Pasten
CERZOS, Bahia Blanca, Argentina
University of Milan (Italy), from May 30, 2024 to August 2, 2024
The objective of this secondment was to initiate the characterization of Arabidopsis thaliana SALK mutant lines previously obtained for the CHC2 (Clathrin heavy chain 2) and NFD3 (Nuclear fusion defective 3) genes, both found to be differentially expressed in apomictic vs sexual genotypes of Eragrostis curvula in previous analyses. Genotyping with specific primers was carried out and several heterozygote lines were found, which are now being studied. Also, we recovered and grew CRISPR-Cas9 mutants for the SPL7 (Squamosa promoter binding like 7) gene. These plants are now under characterization. In addition, in situ hybridizations (ISH) have been carried out to determine the expression profile of the SPL7 gene in the reproductive tissues of diploid and tetraploid individuals in A. thaliana (ecotype Col0).

Domenico Loperfido, MSc
University of Milan, Italy
CERZOS (Bahia Blanca, Argentina) from March 11, 2024 to June 8, 2024
During the three months (from May 11 to June 8, 2024) he spent at CERZOS, Domenico Loperfido worked in the lab of Prof. Echenique under the supervision of José Carballo and Cristian Gallo from CERZOS and that of Prof. Ezquer from the University of Milan. The objective of the secondment was to investigate candidate genes involved in growth and seed development through association studies aiming at understanding the role these genes may have in the inheritance of apomictic processes. Genomes of different Arabidopsis thaliana accessions were used ; using bioinformatics tools, eg bcftools, vcftools, R, GAPIT, and others, significant SNPs in genes phenotypically associated with the growth and the development of seed components (embryo, endosperm, and seed coat) controlling hybrid vigorous in sexual plants were identified

Dr. Diego Zappacosta
CERZOS, Bahia Blanca, Argentina
University of Perugia (Italy) from March 05, 2024 to April 05, 2024
The main objective of this secondment is to continue the characterization of a mapping population of Eragrostis curvula that segregates for apomixis by creating a linkage map and selecting individuals with similar molecular markers but different reproductive modes (apomictic and sexual) for future transcriptomic analysis. During these two months, in collaboration with the group of UNIPG, it is proposed to continue the analysis of the phenotypic and genetic data of a mapping population of E. curvula to generate a new consensus map and to identify molecular markers linked to apomeiosis and, ultimately, to discover genes, which may play a potential role in the control of apomictic reproduction. Additionally, I will work in a calendar to associate floral morphological parameters with megasporogenesis and megagametogenesis developmental stages in different apomictic and sexual genotypes of E. curvula (Task 3.3).
Lucia Giordano, PhD student
University of Perugia, Italy
University of Adelaide (Austalia) from January 18, 2024 to June 2, 2024
2023
Samela Draga, PhD student
University of Padova, Italy
IICAR (Rosario, Argentina) from November 3, 2023 to , January 19, 2024
Samela Draga is visiting Prof. Pessino laboratory at IICAR-CONICET-UNR from November 3, 2023, to January 18, 2024. The general objective of her work is to continue the project already started at March-July 2023, at IICAR-CONIC ET-UNR, contributing to the elucidation of the role of the splicing controller BUD13 (a TGS1 interactor) in apomictic and sexual Paspalum. BUD13 displays differential expression in florets of sexual and apomictic plants in at least two aposporous species, Paspalum notatum and Hypericum perforatum. Interestingly, Arabidopsis bud13 mutants were associated with defects in early embryo development. During her visit, she will generate digoxigenin-labeled BUD13 probe, include Paspalum ovaries in paraffin, slice the blocks, arrange the slices on glass slides and perform the ISH analysis. Observations will allow to determine spatial expression patterns of BUD13 in reproductive organs of sexual and apomictic genotypes of Paspalum notatum.
Dr. Giovanni Gabelli
University of Padova, Italy
IICAR (Rosario, Argentina) from November 15, 2023 to December 15, 2023

Dr. Fulvio Pupilli
IBBR, Perugia, Italy
IICAR (Rosario, Argentina) from November 8, 2023 to December 9, 2023
During this first secondment in Rosario from November 9th to December 14th 2023, Fulvio Pupilli discussed with Juan Pablo Ortiz, his staff and Olivier Leblanc the recent achievements on Nanopore sequencing and on the assembly of apomictic and sexual Paspalum simplex genomes. Furthermore he reported the insights toward the characterization of the gene PsARF5 that is differentially expressed in LCM dissected nucellus tissues of apomictic and sexual genotypes of the same species and discussed with Juan Pablo Ortiz his possible interaction with PnIAA30, a gene under characterization at IICAR. A series of in situ hybridisation experiments for this gene was scheduled for the next secondment. Finally, he prepared a talk on the role of the gene PsORC3 on maternal excess endosperm development in P.simplex and a tribute to Prof. Camilo Luis Quarin for the IV Apomixis Congress 3-7th December 2023 Rosario, Argentina.

Giacomo Bongiorno, PhD student
University of Perugia, Italy
CERZOS (Bahia Blanca, Argentina) from May 18, 2023 to December 09, 2023
During this secondement at CERZOS, my primary focus was on the diplosporous grass Eragrostis curvula, aiming to construct a comprehensive pan-genome. The utilization of the diploid sexual variety, cv. Victoria, as the reference genome provided a solid foundation for investigating genomic variation, phylogenetic relationships, and the impact of ploidy on genome evolution and reproductive modes. This pan-genomic study significantly contributes to our understanding of E. curvula‘s genomic diversity, serving as a valuable resource for targeted investigations into the molecular mechanisms governing its reproductive modes and relationships with ploidy.
Picture (from left to rigth: Juan Pablo Selva, Diego Zappacosta, Juan Manuel Rodrigo, Nengi Lawson, Viviana Echenique, Alejandra Diaz & Giacomo Bongiorno

Dr. Daphné Autran
IRD, Montpellier, France
IICAR (Rosario, Argentina) from October 22, 2023 to December 17, 2023
During this secondment, we follow up the work on Paspalum rufum with Luciana Delgado. To understand the interelations between ovule primordia architecture and cell fate plasticity leading to apospory , we are performing a 3D cellular morphometric analyses of P.rufum genotypes showing differential expressivity of apospory. The first step is to build a reference 3D atlas of the sexual genotype. After the work done in previous secondments of segmentation, labelling, and data export using MorphographX (MGX) software, we set up for quantitative analysis on a pilot group of 11 ovules at successive stages covering the developmental window of interest. In addition to cell number and cell size quantification, different shape descriptors, at both cellular and organ level, were selected and calculated from features extracted by MGX. Since the pilot data already encompass thousands of cells to analyze, custom scripts in R were written to manage data compilation, visualization and statistical analysis. In addition, P.rufum ovules around meiosis stage were dissected, fixed and cleared; to extend the developmental window covered, and maximize the probability of detection of differential cell growth between sexual and aposporous genotypes.
Paspalum rufum, Luciana Delgado & Daphné Autran

Dr. Olivier Leblanc
IRD, Montpellier, France
IICAR (Rosario, Argentina) from October 15, 2023 to December 13, 2023
This second visit to IICAR is part of a secondment dedicated to the genomics of apomixis (WP3) and the functional characterization of Paspalum candidate genes relevant for apomixis using Arabidopsis mutants (WP4). Regarding the genomic approaches, we expect to complete a first draft on the assembly and the detailed annotation of a P. notatum diploid genome reference and will focus analyses on understanding the variations between the genome of diploid, sexual plants and that of apomicts using comparative genomics and k-mer approaches. Regarding the functional characterization of Arabidopsis mutants, the effets of tgs1 mutations on RNA splicing are being analyzed using comparative transcriptional analyses and cytological tools (fluorescent reporters under construction). In addition to these activities, I’m involved in organizing the second MAD workshop on ONT sequencing and bioinformatics (Rosario, November 20-25) and the Apomixis2023 conference (Rosario, December 3-7).
Picture: visiting old friends in Cayasta (Santa Fe, Argentina), cradle of the Paspalum family. from left to right: O Leblanc, JPA Ortiz, & L Siena

Davide Perrone
University of Milan, Italy
IICAR (Rosario, Argentina) from September 12, 2022 to December 13, 2023
Davide Perrone is visiting IICAR-CONICET in order to continue the experiments initiated during a previous secondment, concerning the transformation of apomictic and sexual Paspalum notatum plants with fluorescent auxin and cytokinin markers. The aim of these experiments is to compare the pattern of hormone distribution in the ovule between sexual and apomictic reproductive types. Moreover, in this particular secondment Davide will also start performing in-situ hybridization experiments aimed at comparing the expression of AUXIN RESPONSE FACTOR 10 (ARF10) in ovules of sexual and apomictic Paspalum notatum biotypes. Finally, in collaboration with IICAR scientists, he will perform miR-seq analyses using raw data previously generated in Milan.
Picture (from left to right): Augusto Villalba (IBONE), Silvina Pessino (IICAR), Maricel Podio (IICAR) and Davide Perrone (Univ. Milan)
Dr. Cristian Gallo
Bahia Blanca, Argentina, CERZOS
University of Perugia (Italy), from September 1st, 2023 to October 30, 2023

Dr. Francesca De Marchis
Perugia, Italia, CNR-IBBR
IICAR (Argentina), from August 11, 2023 to September 12, 2023
Francesca De Marchis is a researcher of the National Research Council, CNR- IBBR – Perugia. During her visit, she will work with Dr. Juan Pablo A. Ortiz (IICAR) and her expertise in plant biotechnology will be exploited to design a vector for Paspalum notatum transformation. Some candidates genes for triggering parthenogenesis, identified by the IICAR’s research group, will be considered for plant transformation by biolistic technology choosing for their over-expression or down-regulation.

Dr. Alejandra Díaz
Bahia Blanca, Argentina, CERZOS
NIAB (UK), from August 1st, 2023 to October 2nd, 2023
For some time now, our CERZOS research group has been collaborating with Dr. Caccamo´s group at NIAB on the identification and the characterization of a genomic region involved in apomixis in Eragrostis curvula (Schrad.) Nees and its genes. Some of these, identified previously by our research group to be involved in apomictic pathways, are under functional characterization since many of them do not have an annotation in other species. Expression of these candidate under constitutive promoters in the rice plant model species is one of the approaches used for their characterization. The aim of this secondment at NIAB was to advance the characterization of one of our candidate genes, mainly to determine whether its overexpression led to any defect in embryo sac development. To achieve this, I was involved in the screening of reproductive tissues from spikelets of independent transgenic rice lines generated at NIAB and overexpressing the gene of interest (WP4). We used Differential Interference Contrast (DIC) microscopy to observe reproductive tissues cleared with a methyl salicylate solution, as this technique enables large-scale observations on meiosis and embryo sac development in intact ovules. Thus, the collaboration between NIAB and CERZOS will allow us to get faster characterization of the candiate genes for apomixis identified in Eragrostis.

Dr. Viviana Echenique
Bahia Blanca, Argentina, CERZOS
NIAB (UK), from August 1st, 2023 to October 2nd, 2023
The objective of my secondment at NIAB was to continue with the work started with Dr. Caccamo´s group aiming at the identification and the characterization of a genomic region and genes involved in apomixis. Here we discussed strategies and designed new experiments. The ongoing work consists on the sequencing and the assembly of the genome of the apomictic cultivar Don Walter (Eragrostis curvula). This genome was sequenced at NIAB using the Oxford nanopore platform with the super-high accuracy plant model protocol and one of the students from CERZOS is working here doing the assembly. We also are involved in a transcriptomics project using RNA-seq data obtained from pistil tissues of apomictic and sexual plants in three different developmental stages. We are looking for differentially expressed genes between plants with different reproductive modes and different developmental stages.
Picture: Viviana at The Eagle, Cambridge (UK)

Sol Vega, PhD student
Rosario, Argentina, IICAR
IRD (France), from July 20, 2022 to September 20, 2022
In this secondment María Sol is visiting IRD to continue with the functional characterization of the IAA16 gene in Arabidopsis thaliana, whose orthologue (IAA30) has been associated with apomixis in Paspalum notatum. The activities will be carried out in the laboratory of Dr. Olivier Leblanc and will include the recovery of double homozygous plants derived from crosses between the iaa16 mutant and; a line carrying the pWOX2-CENH3-GFP gametophytic reporter; tgs1 plants (related to alterations in gametophyte development), and; topless plants (involved in the auxin response during embryogenesis). Phenotypic analyses of the reproductive development of the materials obtained will be carried out using fluorescence or DIC microscopy during the secondment and at IICAR. In addition, the study of mutants for other genes from the IAA16 clade (iaa14 and iaa17) as well as for a new IAA16 insertion will be initiated.

Dr. Lorena Siena
Rosario, Argentina, IICAR
IRD (France), from June 22, 2023 to September 16, 2023
In this secondment, Lorena Siena from IICAR, is visiting IRD for three months. The objective of the visit is to advance the functional characterization of the TGS1 and QGJ genes during Arabidopsis thaliana reproductive development. The activities will be carried out in collaboration with the IRD group and, first, include the exploring of the potential for parthenogenetic embryos in tgs1 mutant lines by using embryo-specific fluorescent reporters transmitted maternally and paternally. In addition, the cellular identity of the extra, functional megaspores observed in qgj ovules will be extensively characterized, e.g. using the pWOX2-CENH3-GFP reporter. Moreover, we will initiate experiments aiming at identifying potential interactions between QGJ and TGS1 by determining QGJ expression pattern (pQGJ-GUS) in tgs1 ovules and, reciprocally, that of TGS1 (pTGS1:TGS1-GUS) in qgj ovules.
Picture: Montpellier aqueduct (@ Lorena Siena, IICAR).

Dr. Jose Carballo
Bahia Blanca, Argentina, CERZOS
NIAB (UK), from June 14, 2023 to September 14, 2023
The objective of my secondment at NIAB was to continue with the sequencing and assembly of the apomictic genome of Eragrostis curvula. The genome of Don Walter genotype was sequenced at NIAB institute using the Oxford nanopore platform with the super-high accuracy plant model protocol. These reads were assembled using Flye software and scaffolded with Omni-C technology in order to obtain a chromosome scale assembly. This genome was assembled and annotated obtaining the A and B sub-genomes of the allotetraploid Don Walter. Moreover, during this time in Cambridge, the RNA-seq data obtained from pistil tissues of apomictic and sexual plants in three different developmental stages was analysed obtaining differentially expressed genes. This secondment continues with the collaborative efforts between NIAB and CERZOS-CONICET in order to disclose the molecular basis of apomixis through a bioinformatic approach.

Dr. Tamunonengiye-ofori Lawson
Cambridge, UK, NIAB
CERZOS (Argentina), from May 20, 2023 to July 19, 2023
The objective of this secondment at CERZOS was to gain deeper knowledge, skills and practical experiencein teh area of in-situ hybridization and DIC microscopy in plants.
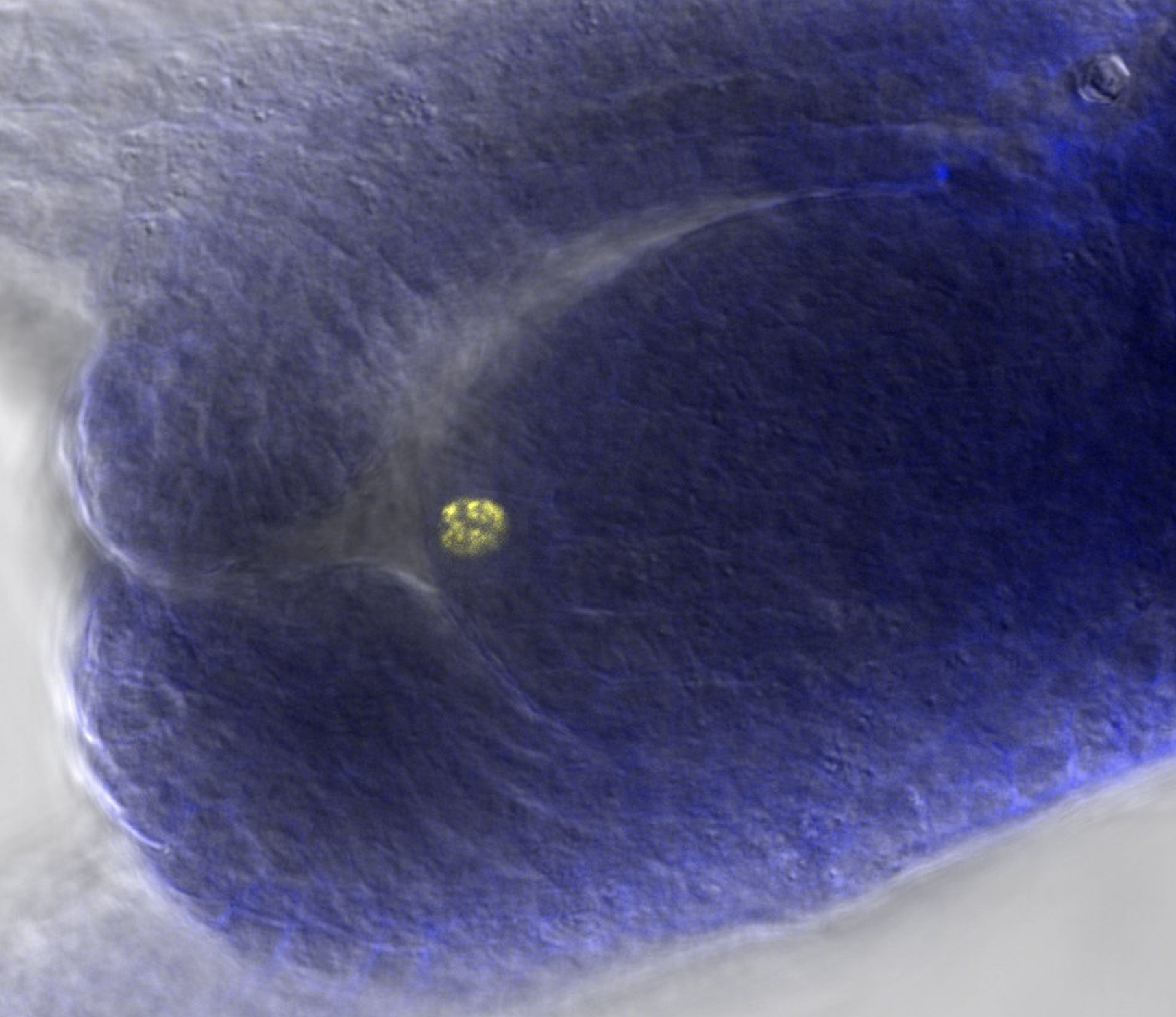
Dr. Luciana Delgado
Rosario, Argentina, IICAR
IRD (France), from May 20, 2023 to August 19, 2023
This secondment continues the tasks initiated in 2022 with Dr. Daphné Autran to build a 3D atlas of early ovule development in Paspalum rufum. We will segment and classify the cells within ovules primordia using high resolution images obtained previously, and a previously established and partially automated workflow using the open source software MorphoGraphX. In parallel, we will test different staining protocols to allow incorporation and detection of a thymidine analogue (EdU) for quantifying location and duration of cell divisions during ovule development in different grasses, including two sexual model species (rice and maize) and Paspalum apomicts. This protocol was successfully used in Arabidopsis and a few other species, it will be useful optimizing conditions for grass species showing disparities in flower and ovule morphologies. Quantitative information from the 3D Atlas and cell division in sexual and apomictic genotypes should help us understand the switch between the two reproductive pathways.
Picture: Interphase cell in a young ovule of Paspalum rufum showing fluorescent signal from DNA containing EdU (@ Luciana Delgado, IICAR).

Samela Draga, PhD student
University of Padova, Italy
IICAR (Rosario, Argentina) from May 16, 2023 to Aug. 2, 2023
The general objective of Samela’s work at IICAR is to contribute to the elucidation of the role of the splicing protein BUD13 in the apomixis-sexuality switch. During her stay she will analyze the representation of BUD13 in transcriptomic and genomic databases of Paspalum notatum and generate suitable RNA hybridization probes. Moreover, she will classify ovules of sexual and apomictic Paspalum notatum plants according to developmental stages by using DIC cytoembryology, dehydrate them in an ethanol/xylol series and include them in paraffin. In a second visit (from October to December) she will prepare ovule sections for hybridization using sense and antisense digoxigenine-labelled RNA probes detected by colorimetric reaction using an anti-DIG conjugated with alkaline phosphase and NBT/BCIP substrates. We expect to produce information on the spatial differential expression of these candidate genes in apomictic and sexual genotypes.
Samela Draga (right) & Silvina Pessino (left)

Nicola Babolin, PhD student
University of Milan, Italy
University of Adelaide (Australia) from April 10, 2023 to October 13, 2023
During my secondment in the University of Adelaide (Australia) in A/Prof. Matthew Tucker laboratory I’m willing to better characterize the molecular network between the two MADS-box genes STK and ABS during the fertilization process and seed development in Arabidopsis thaliana.
At the same time I will be involved on the projects run by Prof. Tucker team on barley plant concerning ovule and seed development highly interconnected with my project in Arabidopsis.
From left to right: Nicola Babolin, Mathew Tucker, Rosanna Petrella

Dr. Rosanna Petrella
University of Milan, Italy
University of Adelaide (Australia) from April 10, 2023 to October 13, 2023
During my secondment at The University of Adelaide (Matthew Tucker’s lab) my work will be focused on unravelling the molecular mechanisms determining a correct female germline progression in Arabidopsis thaliana. In particular, I will try to characterize the main players involved in spore degeneration and functional megaspore selection and differentiation at the end of megasporogenesis in the ovule. I will determine the contribution of phytohormone cross-talk by trying to rescue mutants defective in this key processes using specific regulatory regions active in different domain of the ovule to specifically drive the expression of candidate genes.
From left to right: Rosanna Petrella, Skippy the Kangaroo, Nicola Babolin

Mattia Spano, PhD student
University of Milan, Italy
LANGEBIO (Mexico) from April 1, 2023 to July 20, 2023
The aim of this secondment is to investigate molecular mechanism determining female germline. In particular, I will investigate the contributions of phytohormones, auxin and cytokinines, in MMC differentiation. The species investigated are Arabidopsis thaliana and Oryza sativa var. Kitaake. In this frame, I will perform Immunolocalization assays in different backgrounds to localize phytohormones during ovule development using spl/nzz mutant and wild type Arabidopsis and wild type for Oryza sativa.

Dr. Juan Pablo A. Ortiz
Rosario, Argentina, IICAR
IRD (France), from March 7, 2023 to April 3rd, 2023
A new stay at IRD of Dr. Juan Pablo A. Ortiz from IICAR, carried out as part of a MAD secondment for the characterization of apomictic genomes (WP3), started March 7, 2023. The work plan includes DNA extraction of several samples of Paspalum spp, intended for Illumina sequencing and further genome structural analyses. The visit is also dedicated to preparing a new manuscript describing the sequencing, chromosome-level assembly and annotation of a Paspalum notatum diploid genome. This achievement is a key step for WP3 progression because it will serve as a reference for characterizing tetraploid genomes and determining the structure and evolution of the apomixis region. This manuscript is the result of a collaborative effort between MAD partners from Argentina, Italy, and France. Finally, the program of the international workshop on Nanopore sequencing and bioinformatics, organized by the MAD project in Rosario, Argentina next November will be discussed and decided during this visit.
IRD plant greenhouse. Juan Pablo A. Ortiz (right) & Olivier Leblanc (left)

Dr. Alejandra Díaz
Bahia Blanca, Argentina, CERZOS
University of Milan (Italy) from Jan 21, 2023 to March 23, 2023
The aim of this secondment was to analyze the tissue-specific expression of two candidate genes involved in the apomictic reproductive mode in the grass Eragrostis curvula (WP4). For this purpose, we used RNA in situ hybridization technic (RNA-ISH) to determine the expression patterns cell-specific of these two genes in the reproductive organs of E. curvula, both in the full apomictic and sexual genotypes. In addition, we also characterized a gene that shows differential expression between sexual and apomictic E. curvula plants and that is also expressed in the model plant A. thaliana. For the latter objective, we also used ISH to test the RNA abundance and localization of this orthologous gene in the reproductive tissues of E. curvula (apomictic and sexual genotypes) and A. thaliana (diploid and tetraploid genotypes).
From left to right: Mareta Mendes, Lucia Colombo, Alejandra Díaz and Cielo Pasten
2022
Gregorio Orozco
University of Milan, Italy
LANGEBIO (Mexico) from Dec. 7, 2022 to Feb. 3, 2023
Dr. Lucia Colombo
University of Milan, Italy
IICAR (Rosario, Argentina) from Nov. 14, 2022 to Nov. 27, 2022
Jimena Gallardo
Bahia Blanca, Argentina, CERZOS
University of Perugia (Italy), from Nov. 2, 2022 to Dec 2, 2022

Dr. Olivier Leblanc
Montpellier, France, IRD
IICAR (Argentina) from Oct. 17, 2022 to Dec. 16, 2022
This secondment aims at pursuing (1) the characterization of the chromosomic region associated with apomixis in tropical forages. As we now have completed the assembly and the annotation of the genome of a diploid, sexual individual, different approaches will be explored to reveal structural genomic variations that underlie the apomictic development in Paspalum species. To achieve this, we produced > 100 Gb of long reads (Nanopore technology) from several Paspalum apomictic races that will be used for de novo assembly and comparative analyses, and; (2) the functional analysis of tgs1 Arabidopsis mutants and, more particularly, the comparative transcriptional analysis in tgs1 and WT Arabidopsis embryos collected at early developmental stages. These activities are part of WP3 and WP4, respectively.
Picture: Metallic clones in the pampa outside of Rosario, Province of Santa Fe, Argentina (@ O Leblanc)

Dr. Maria Cielo Pasten
Bahia Blanca, Argentina, CERZOS
University of Milan (Italy) from Sept. 30, 2022 to March 1, 2023
The objective of this detachment was to analyze genes that show differential expression between sexual and apomictic ovules of Eragrostis curvula plants and, which are also present in the model plant Arabidopsis thaliana. For this purpose, we studied the phenotype of A. thaliana mutants publicly available for some of these genes but, also, that of new transgenic mutant lines generated by the CRISPR-Cas9 strategy. In addition, ovule development in apomictic and sexual genotypes of E. curvula was characterized using confocal microscopy to further our knowledge on the structures and differences between sexual and asexual modes of reproduction.
From left to right: Marta Mendes, Lucia Colombo, Alejandra Díaz and Cielo Pasten

Pr. Pablo Roncallo
Bahia Blanca, Argentina, CERZOS
University of Perugia (Italy) from Sept. 6, 2022 to Nov. 8, 2022
The objective of this secondment is to extend a tetraploid genetic linkage map of Eragrostis curvula using DarT seq markers and additional F1 genotypes in order to map more precisely the region involved in apomixis. The F1 population was obtained by crossing a sexual (OTA) and an apomictic genotypes (Don Walter) as maternal and paternal parents, respectively. This new linkage map will facilitate the localization of loci controlling apomixis (diplospory) and to study the genome evolution in this grass (WP3). In collaboration with the UNIPG group, I am analyzing molecular markers (DarT seq) obtained in new F1 individuals and in others previously obtained in Bahía Blanca by our group to construct a new integrated linkage map, as well as two parent-specific linkage maps. For this, segregation analysis and detection of polymorphic SNP markers in one or both parent is needed to solve this three linkage maps. Additionally, the haplotype reconstruction using this F1 population will be explores and parental SNP profile.

Davide Perrone
University of Milan, Italy
IICAR (Rosario, Argentina) from Sept. 2, 2022 to Nov. 28, 2022
Davide Perrone (University of Milan, Italy) is visiting Prof. Silvina Pessino laboratory at IICAR CONICET, Rosario, Argentina, from 3rd September to 27th November 2022. His work, conducted in collaboration with Dr. Carolina Colono (IICAR-CONICET-UNR), is aimed at using biolistic transformation to produce DR5 (auxin marker) and TCS (cytokinin marker) Paspalum notatum fluorescent lines with different reproductive modes (sexuality and aposporous apomixis). These plant materials will be useful to study the ovular hormone patterns along different developmental stages in a well-characterized apomictic model. Moreover, Davide is contributing to study the role of AUXIN RESPONSE FACTOR 10 (ARF10) in the transport of auxin within the Arabidopsis ovule, by doing cytoembryological characterizations of arf10 x PIN1 and arf10 x DII-Venus crosses previously obtained at IICAR-CONICET-UNR. Finally, he is involved in the analysis of the functional connexion between AUXIN RESPONSE FACTOR 10 (ARF10) and TRIMETHYL GUANOSINE SYNTHASE (TGS1) (a reported apospory controller) in Paspalum.
From left to rigth: Carolina Colono, Silvina Pessino and Davide Perrone

Sol Vega, PhD student
Rosario, Argentina, IICAR
IRD (France), from June 15, 2022 to August 16, 2022
Maria Sol is a PhD fellow at IICAR-CONICET, Argentina, and she is visiting the laboratory of Dr. Olivier Leblanc at IRD (Montpellier, France). The main objective of her stay is to initiate the functional analysis of Arabidopsis mutants defective for the IAA16 gene, whose orthologue in Paspalum notatum (PsIAA30) is down-regulated during apomictic development (Siena et al., Plants 2022, 11, 472). Heterozygous and homozygous iaa16 mutants will be analyzed by cytoembryological observations to search for possible female reproductive phenotypes reminiscent of aposporous apomixis. These activities will then contribute to determine the effects of IAA16 depletion on the reproductive development of Arabidopsis and to plan further functional characterization for this or other genes that belong to the pathway. During her visit, Maria Sol will work in collaboration with Dr. Olivier Leblanc and Caroline Michaud from IRD, and with Lorena Siena, her PhD supervisor at IICAR.
Lorena Siena (left) & Sol Vega (right)

Dr. Juan Pablo A. Ortiz
Rosario, Argentina, IICAR
IRD (France), from June 7, 2022 to July 7, 2022
A new MAD secondment to work on the characterization of apomictic genomes (WP3) started on June 7, 2022. On this occasion, Dr. Juan Pablo A. Ortiz from IICAR (CONICET-UNR, Argentina) is visiting the laboratory of Dr. Olivier Leblanc at IRD (France). The workplan includes the annotation of a genome assembly of a diploid Pensacola bahiagrass ecotype (Paspalum notatum, var. saurae Parodi), obtained previously in a collaboration between laboratories of France, Italy, and Argentina. We also intend to identify and to characterize sequences related to the apomixis controlling locus (ACL) in a tetraploid race. In particular, we will use the ONT “adaptive sampling” technology that allows sequencing of regions of interest. Therefore, we expect to enrich our sequencing with reads specific from the tetraploid apomictic plant and then assemble the ACL. Olivier Leblanc, Cédric Mariac and Julie Orjuela (DIADE research Unit), as well as Juan Manuel Vega (MAD seconded from IICAR) will contribute to this work.
Juan Pablo Ortiz (left) & Olivier Leblanc (right)

Dra. Lorena Siena
Rosario, Argentina, IICAR
IRD (France), from June 6, 2022 to September 1st, 2022
During this 3-month secondment, Lorena Siena (IICAR, CONICET- Argentina) is visiting IRD with the objective of advancing the functional characterization of the TGS1 gene during the reproductive development of the model plant Arabidopsis thaliana. The activities will be carried out in collaboration with Dr. Olivier Leblanc and his IRD group, and include: complementation tests using the pTGS1:TGS1-GUS construct; determining the origin of the extra embryo sacs observed in tgs1 mutants using reporter lines carrying the pWOX2 and CENH3 genes reporting both female gametophytic identity and chromosome number (reduced vs. unreduced origin) , and; analysis of RNAseq data obtained from wild type and tgs1 embryos at early stages of embryonic development. This study aims to determine the alterations in gene expression patterns caused by TGS1 depletion; to identify their potential target genes, and; to explore whether alternative splicing controls early embryogenesis in plants.
Olivier Leblanc (left) & Lorena Siena (right)
Dra. Luciana Delgado
Rosario, Argentina, IICAR
IBBR (Italy), from May 5, 2022 to June 6, 2022
In this secondment, Dr Luciana Delgado (IICAR) and Dr Daphné Autran work together to study the communication between cell division, expansion and differentiation throughout female reproductive development in P. rufum, an apomictic species. The overall aim is to build a 3D atlas of early ovule development. During the stay we will establish the optimal conditions for clarification of P. rufum ovules and cell wall staining techniques to obtain high-resolution 3D images. The images will be segmented with MorphoGraphX, classified by developmental stage, and finally a quantitative analysis of cellular descriptors will be performed. We believe that understanding the coordination between reproductive developmental processes with cell growth and division, in an apomictic context could be enlightening for understanding the regulation of apomixis.
Daphné Autran (left) & Luciana Delgado (right)

Dr. Juan Pablo Selva
Bahia Blanca, Argentina, CERZOS
University of Milan (Italy), from May 16, 2022 to July 16, 2022
The objective of this secondment was to continue with the functional characterization of a gene differentially expressed in sexual and apomictic plants of Eragrostis curvula using maize mutants carrying a Mutator insertion in the orthologous gene.
During these two months, I collaborated with the group of Professor Gabriella Consonni to evaluate different maize F2 populations that were planted in an experimental field. Plants were genotyped using specific primers for the transposon insertion located in the first exon of the orthologous gene and specific primers for the wild type allele. Activities also included observations of cleared pistils under the microscope searching for abnormalities during female reproductive development.
Dr. Juan Pablo A. Ortiz
Rosario, Argentina, IICAR
IBBR, from May 5, 2022 to June 6, 2022
A new MAD secondment involving researchers from Argentina and Italy started May 9, 2022 for one month! Indeed, Dr. Juan Pablo A. Ortiz from IICAR, (CONICET-UNR) is visiting Dr. Fulvio Pupilli’s lab at the Institute of Biosciences and Bioresources (IBBR-CNR) of Perugia. The working plan is focused on the functional analysis of ORC3, a gene that segregates as completely linked to apomixis, in the formation of the endosperm of Paspalum simplex apomicts. Moreover, it is also intented to progress in the assembly and the characterization of the P. simplex genome using long reads generated using the Oxford Nanopore sequencing technology. Dr. Pupilli group’s members involved in these projects are: Dra. María Eugenia Cáceres, Dr. Andrea Rubini, Dra. Francesca De Marchis and Dr. Michele Bellucci. By the end of the secondment, we expect to have completed the redaction of a manuscript on PsORC3 functional analysis and produced a draft assembly of the P. simplex genome.
Fulvio Pupilli (left) & Juan Pablo A. Ortiz (right)

Dr. Carolina Colono & Prof. Silvina Pessino
Rosario, Argentina, IICAR
University of Milan, from April 29, 2022 to July 2nd, 2022
From April 29 to July 2nd, 2022 Dr. Carolina Colono and Prof. Silvina Pessino will be visiting Prof. Lucia Colombo laboratory at the University of Milan, to explore the role of the hormonal control in the transition from sexuality to apomixis using the model plant Arabidopsis thaliana.
From left to rigth: Carolina Colono, SIlvina Pessino and Lucia Colombo

Juan Manuel Vega, PhD student
Rosario, Argentina, IICAR
IRD, Montpellier (France), from April 19, 2022 to June 18, 2022
This secondment is part of the effort undertaken by the MAD project (see WP3 objectives) to determine the nature, the evolution and the functional role of the apospory controlling region (ACR) in Paspalum notatum. By taking advantage of a diploid reference genome and long reads data sets available for tetraploid apomicts, I will first use the adaptive sampling method offered by the ONT sequencing platform to enrich sequencing of ACR long reads and then perform a high coverage assembly for both an obligate apomict and a facultative apomict. Complementary approaches by using open source methods aiming at identifying and filtering structural variations will also be applied. Altogether, this should allow structural and comparative genomics to determine the genetic content and coding/regulatory capacity within the ACR and provide cues for its functional role in the emergence of apomixis in Paspalum.

Dr. Diego Zappacosta
Bahia Blanca, Argentina, CERZOS
University of Perugia (Italy), from March 1st, 2022 to May 7, 2022
The objective of this secondment is to continue the assembly of Eragrostis curvula tetraploid genomes to determine the locus controlling apomixis and study genome evolution in this grass (WP 3). During these two months, I will analyze, in collaboration with the group of UNIPG, the genetic relationships between the sexual diploid (cv Victoria), previously sequenced and published by the 2 institutions, and the apomictic tetraploid cv. Tanganyika. We will first assemble a pan-genome and resolve it into the core genome and the dispensable genome. The reads mapping onto the Victoria reference will be assembled while unmapped reads will be assembled de novo to analyze the presence/absence variations. We will next analyze orthologous genes to find contractions and expansions of gene families and compare regions controlling apomixis in Eragrostis and Paspalum spp. Finally, I will apply the MCSeEd protocol developed by UNIPG to compare differentially methylated regions between apomictic and sexual genotypes (Task 3.3).
